# packages
library(galah)
library(ggplot2)
library(dplyr)
library(sf)
library(ozmaps)
library(tidyr)
library(stringr)
galah_config(email = "your-email-here") # ALA-registered email
desert_plant <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.25ba5f73-3fdc-4ad3-a702-ada03e605de0") |>
atlas_occurrences()
frogs <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.0ede6a98-4c01-461c-ab89-5bce002cdd84") |>
atlas_occurrences()
native_mice <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.bf20ceb6-4482-4bfb-99f9-8908da0fe3e0") |>
atlas_occurrences()
acacias <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.9b678ec0-560b-4837-b2bc-d790dcbaf67e") |>
atlas_occurrences()
butterflies <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.f55459dc-441e-4fd3-9f46-ceeb031aa656") |>
atlas_occurrences()
bitter_peas <- galah_call() |>
filter(doi == "https://doi.org/10.26197/ala.706613ae-ee17-471b-90c1-c9fb87830fe4") |>
atlas_occurrences()10 Geospatial cleaning
Geospatial observational data provides essential information about species’ locations over time and space, and can be used with ecological data to understand interactions of species with their environment. However, geospatial data can be difficult to work with. Seemingly minor issues can have large impacts on the data’s validity.
Outliers—points located at large-enough distances from the majority of the species’ observations to skew an overall distribution—are a major challenge for geospatial data cleaning. Identifying outliers can be difficult because it’s not always clear whether an outlier is a true outlier or a data error. Data errors can result from species misidenfitication or incorrect geo-referencing. Other accidental data errors like reversing numeric symbols, mistyping a coordinate number or entering the wrong location can have dramatic consequences on a species’ reported location as well. For some species with smaller ranges, these errors will be easier to find. For species with larger ranges or analyses with many species over a larger area, these errors will be much more difficult to identify.
Every dataset has its own combination of issues that will require bespoke cleaning to address them. It is important to clean geospatial data effectively if they are to be useful, as geospatial errors can lead to unexpected results of species range estimates and analytic output.
In this chapter, we show some common issues with coordinate data and how to correct or remove records that appear suspicious.
10.0.1 Prerequisites
In this chapter we’ll use several datasets:
- MacDonnell’s desert fuschia (Eremophila macdonnellii) occurrence records from the ALA
- Red-eyed tree frog (Litoria chloris) occurrence records in 2013 from the ALA
- Kowari (Dasyuroides byrnei, a native mouse) occurrence records from the ALA
- Acacia occurrence records from the ALA
- Common brown butterfly (Heteronympha merope) occurrence records in 2014 from the ALA
- Bitter pea (Daviesia ulicifolia) occurrence records from the ALA
10.1 Missing coordinates
Mentioned in the Missing Values chapter, many spatial analytical tools are not compatible with missing coordinate data. We recommend identifying the rows that have missing data before deciding to exclude them.
# Identify missing data in coordinates
desert_plant |>
filter(is.na(decimalLatitude) | is.na (decimalLongitude))# A tibble: 74 × 9
recordID scientificName taxonConceptID decimalLatitude decimalLongitude
<chr> <chr> <chr> <dbl> <dbl>
1 000d4874-8c74… Eremophila ma… https://id.bi… NA NA
2 050653b6-a41c… Eremophila ma… https://id.bi… NA NA
3 06e69581-7d6d… Eremophila ma… https://id.bi… NA NA
4 0eead38f-0c16… Eremophila ma… https://id.bi… NA NA
5 0f52e34b-a803… Eremophila ma… https://id.bi… NA NA
6 1190b3b9-90d8… Eremophila ma… https://id.bi… NA NA
7 18d11ae3-e558… Eremophila ma… https://id.bi… NA NA
8 19426c52-9d49… Eremophila ma… https://id.bi… NA NA
9 205d432e-c6bc… Eremophila ma… https://id.bi… NA NA
10 2ab6846b-00cb… Eremophila ma… https://id.bi… NA NA
# ℹ 64 more rows
# ℹ 4 more variables: eventDate <dttm>, occurrenceStatus <chr>,
# dataResourceName <chr>, PRESUMED_SWAPPED_COORDINATE <lgl>You can use drop_na() to remove missing values from your dataset.
# Excluding them
desert_plant <- desert_plant |>
tidyr::drop_na(decimalLatitude, decimalLongitude)- 1
-
You could also use
drop_na(starts_with("decimal"))to achieve the same thing
10.2 Issues to fix records
Spatial outliers can sometimes be due to taxonomic misidentification, but not always. Sometimes, records that appear as outliers can be true observations of a species, but the record has a mistake in its coordinates. To avoid deleting data that can be included in your analysis, it’s good practice to use several sources of spatial information to decide whether an unexpected data point is due to a small but fixable error in coordinates, or not.
Many coordinates issues can be solved with data manipulation instead of discarding. Here are several coordinate issues that can be identified and corrected. Follow the link to each case study to learn how to identify and fix the issue.
10.2.1 Swapped numeric sign
If there is a clustering of points mirrored to another hemisphere, consider swapping the sign and correct rather than discarding the points.
Let’s use MacDonnell’s desert fuschia occurrence records for our example (downloaded at the start of the chapter). If using the galah package, we can add the PRESUMED_SWAPPED_COORDINATE assertion column to our download to help find occurrence records flagged as suspicious for swapped coordinates.
desert_plant <- desert_plant |>
drop_na(decimalLongitude, decimalLatitude) # remove NA coordinates
desert_plant |>
select(PRESUMED_SWAPPED_COORDINATE, everything())# A tibble: 889 × 9
PRESUMED_SWAPPED_COO…¹ recordID scientificName taxonConceptID decimalLatitude
<lgl> <chr> <chr> <chr> <dbl>
1 FALSE 0009009… Eremophila ma… https://id.bi… -22.8
2 FALSE 002e372… Eremophila ma… https://id.bi… -25.3
3 FALSE 0034dc0… Eremophila ma… https://id.bi… -23.9
4 FALSE 0063223… Eremophila ma… https://id.bi… -24.5
5 FALSE 00d15a5… Eremophila ma… https://id.bi… -27.8
6 FALSE 013049a… Eremophila ma… https://id.bi… -25.2
7 FALSE 015571a… Eremophila ma… https://id.bi… -22.1
8 FALSE 01b5e44… Eremophila ma… https://id.bi… -25.2
9 FALSE 02524e1… Eremophila ma… https://id.bi… -25.1
10 FALSE 026c225… Eremophila ma… https://id.bi… -25.8
# ℹ 879 more rows
# ℹ abbreviated name: ¹PRESUMED_SWAPPED_COORDINATE
# ℹ 4 more variables: decimalLongitude <dbl>, eventDate <dttm>,
# occurrenceStatus <chr>, dataResourceName <chr>We can see this single record highlighted in orange on our map, sitting in a very similar location to where Australia would be if we mirrored its location.
# Retrieve map of Australia
aus <- st_transform(ozmap_country, 4326)
ggplot() +
geom_sf(data = aus) +
geom_point(data = desert_plant,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = PRESUMED_SWAPPED_COORDINATE)) +
pilot::scale_color_pilot()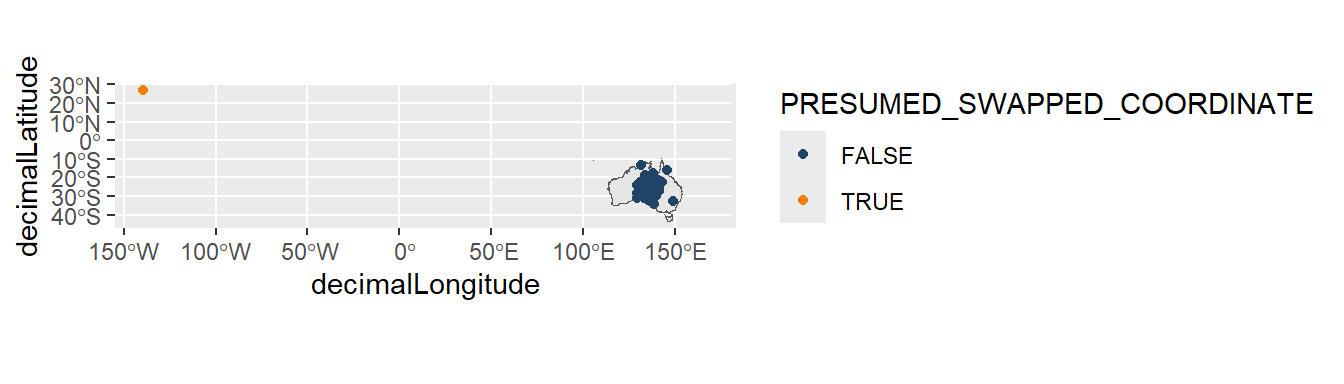
We can fix the numeric symbols using case_when() from dplyr, which works the same as an ifelse statement (but can handle many of statements at once). The first updates our decimalLongitude column so that when decimalLongitude is less than 0, we remove the negative symbol by multiplying by -1, otherwise we keep the original longitude value. The second updates our decimalLatitude column using the same process.
desert_plant_filtered <- desert_plant |>
mutate(
decimalLongitude = case_when(
decimalLongitude < 0 ~ decimalLongitude * -1,
.default = decimalLongitude
),
decimalLatitude = case_when(
decimalLatitude > 0 ~ decimalLatitude * -1,
.default = decimalLatitude
))Our updated map has fixed the coordinates of our record.
ggplot() +
geom_sf(data = aus) +
geom_point(data = desert_plant_filtered,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = PRESUMED_SWAPPED_COORDINATE)) +
pilot::scale_color_pilot()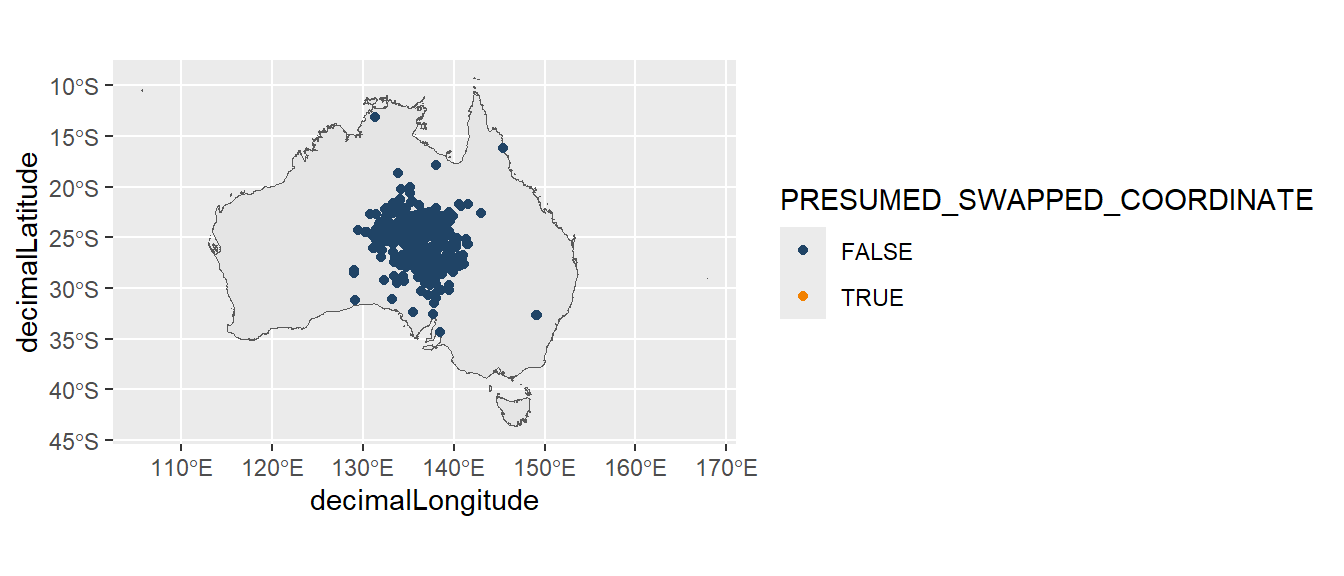
10.2.2 Location description doesn’t match coordinates
Sometimes not all of the metadata about location aligns with the coordinate location. Occasionally these errors are fixable. Let’s use red-eyed tree frog data as an example (downloaded at the start of the chapter).
frogs <- frogs |>
drop_na(decimalLatitude, decimalLongitude) # remove NA values
frogs# A tibble: 30 × 14
recordID scientificName taxonConceptID decimalLatitude decimalLongitude
<chr> <chr> <chr> <dbl> <dbl>
1 0bbcdc4a-5638… Litoria chlor… https://biodi… -28.4 153.
2 16ecde94-6b9b… Litoria chlor… https://biodi… -28.4 153.
3 22b115c9-f799… Litoria chlor… https://biodi… -28.2 153.
4 236bda61-799f… Litoria chlor… https://biodi… -20.3 149.
5 2ba9c818-e81a… Litoria chlor… https://biodi… -27.4 152.
6 4a1b77fa-3538… Litoria chlor… https://biodi… -30.1 153.
7 4d3274a4-f9cd… Litoria chlor… https://biodi… -27.4 153.
8 4e87c52b-2b80… Litoria chlor… https://biodi… -28.2 153.
9 52c1043c-5f79… Litoria chlor… https://biodi… -29.7 152.
10 52d5551a-816d… Litoria chlor… https://biodi… -29.9 153.
# ℹ 20 more rows
# ℹ 9 more variables: eventDate <dttm>, occurrenceStatus <chr>,
# dataResourceName <chr>, countryCode <chr>, locality <chr>, family <chr>,
# genus <chr>, species <chr>, cl22 <chr>When we plot the coordinates of our red-eyed tree frog occurrences, there is an unexpected observation near Japan. This is quite surprising—red-eyed tree frogs are not native to Japan!
# Get a map of aus, transform projection
aus <- ozmaps::ozmap_country |>
st_transform(crs = st_crs(4326))
# Map
ggplot() +
geom_sf(data = aus,
fill = NA,
colour = "grey60") +
geom_point(data = frogs,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = "#557755")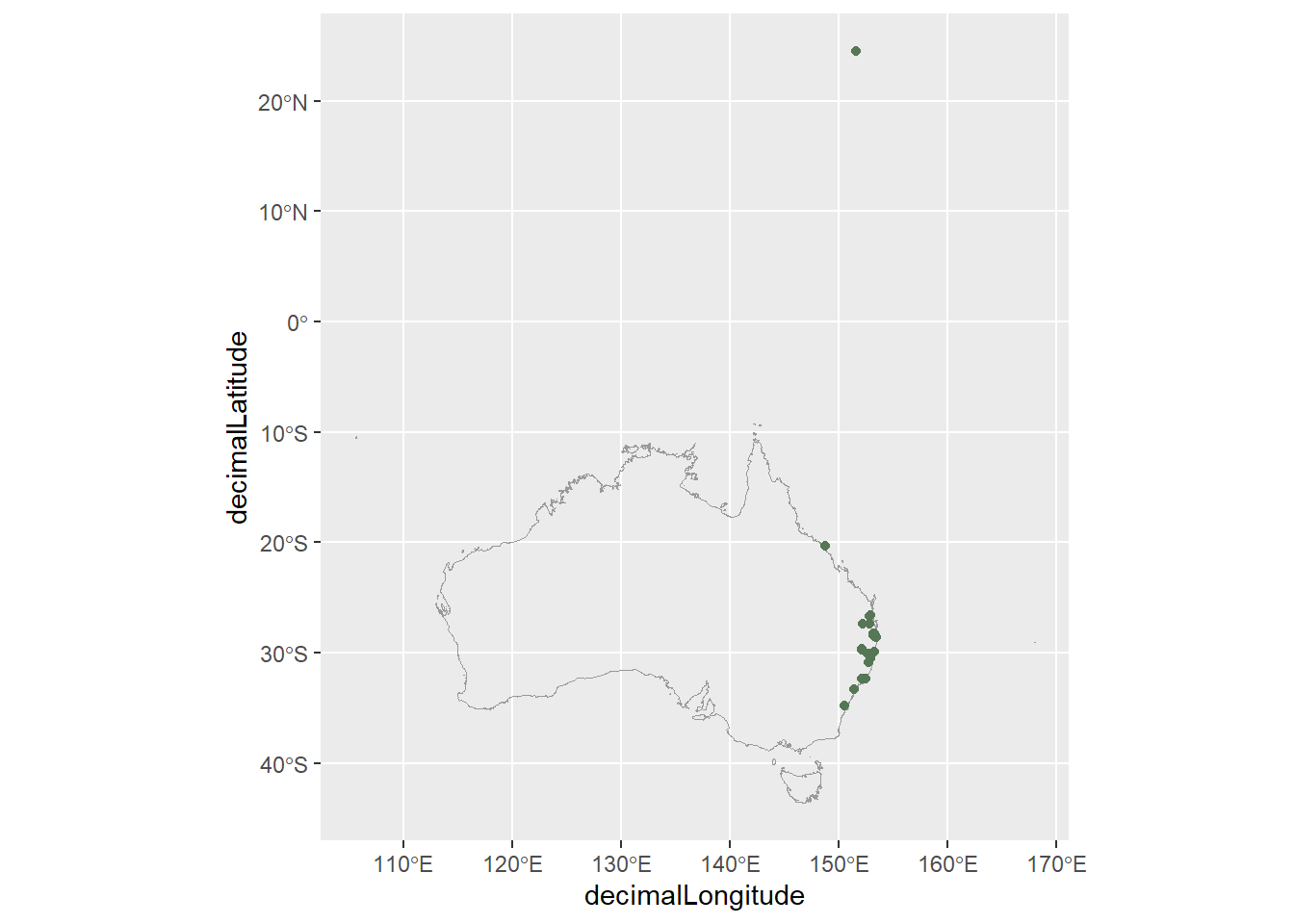
Let’s check the countryCode column to see whether this might be an Australian record with a mistake in the coordinates. Using distinct(), we can see that there are 2 country codes…
frogs |>
distinct(countryCode)# A tibble: 2 × 1
countryCode
<chr>
1 AU
2 JP …and filtering to Japan ("JP") identifies our stray data point.
frogs |>
filter(countryCode == "JP")# A tibble: 1 × 14
recordID scientificName taxonConceptID decimalLatitude decimalLongitude
<chr> <chr> <chr> <dbl> <dbl>
1 c08e641e-cf01-… Litoria chlor… https://biodi… 24.5 152.
# ℹ 9 more variables: eventDate <dttm>, occurrenceStatus <chr>,
# dataResourceName <chr>, countryCode <chr>, locality <chr>, family <chr>,
# genus <chr>, species <chr>, cl22 <chr>So far this observation does seem to be in Japan. To be extra certain, we can also use the column locality, which provides additional information from the data collector about the record’s location.
frogs |>
filter(countryCode == "JP") |>
select(countryCode, locality, scientificName, decimalLatitude, decimalLongitude)# A tibble: 1 × 5
countryCode locality scientificName decimalLatitude decimalLongitude
<chr> <chr> <chr> <dbl> <dbl>
1 JP mt bucca Litoria chloris 24.5 152.The locality column reveals the observation was made in “mt bucca”. This is surprising to see because Mt Bucca is a mountain in Queensland!
When we look at our Japan data point’s decimalLongitude and decimalLatitude alongside other values in our data, it becomes clear that the Japan data point seems to sit within the same numerical range as other points, but the decimalLatitude is positive rather than negative.
frogs |>
arrange(desc(countryCode)) |>
select(countryCode, decimalLongitude, decimalLatitude) |>
print(n = 5)# A tibble: 30 × 3
countryCode decimalLongitude decimalLatitude
<chr> <dbl> <dbl>
1 JP 152. 24.5
2 AU 153. -28.4
3 AU 153. -28.4
4 AU 153. -28.2
5 AU 149. -20.3
# ℹ 25 more rowsAll of this evidence suggests that our Japan “outlier” might instead be an occurrence point with a mis-entered latitude coordinate.
Let’s fix this by adding a negative symbol (-) to the record’s latitude coordinate number. We’ll use case_when() from dplyr to specify that if the countryCode == "JP", then we’ll multiply the decimalLatitude by -1, reversing the symbol.
frogs_fixed <- frogs |>
mutate(
decimalLatitude = case_when(
countryCode == "JP" ~ decimalLatitude * -1,
.default = decimalLatitude
))
frogs_fixed |>
filter(countryCode == "JP") |>
select(decimalLatitude, decimalLongitude, countryCode)# A tibble: 1 × 3
decimalLatitude decimalLongitude countryCode
<dbl> <dbl> <chr>
1 -24.5 152. JP Mapping our data again shows our outlier is an outlier no longer!
Code
ggplot() +
geom_sf(data = aus,
fill = NA,
colour = "grey60") +
geom_point(data = frogs_fixed,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = "#557755")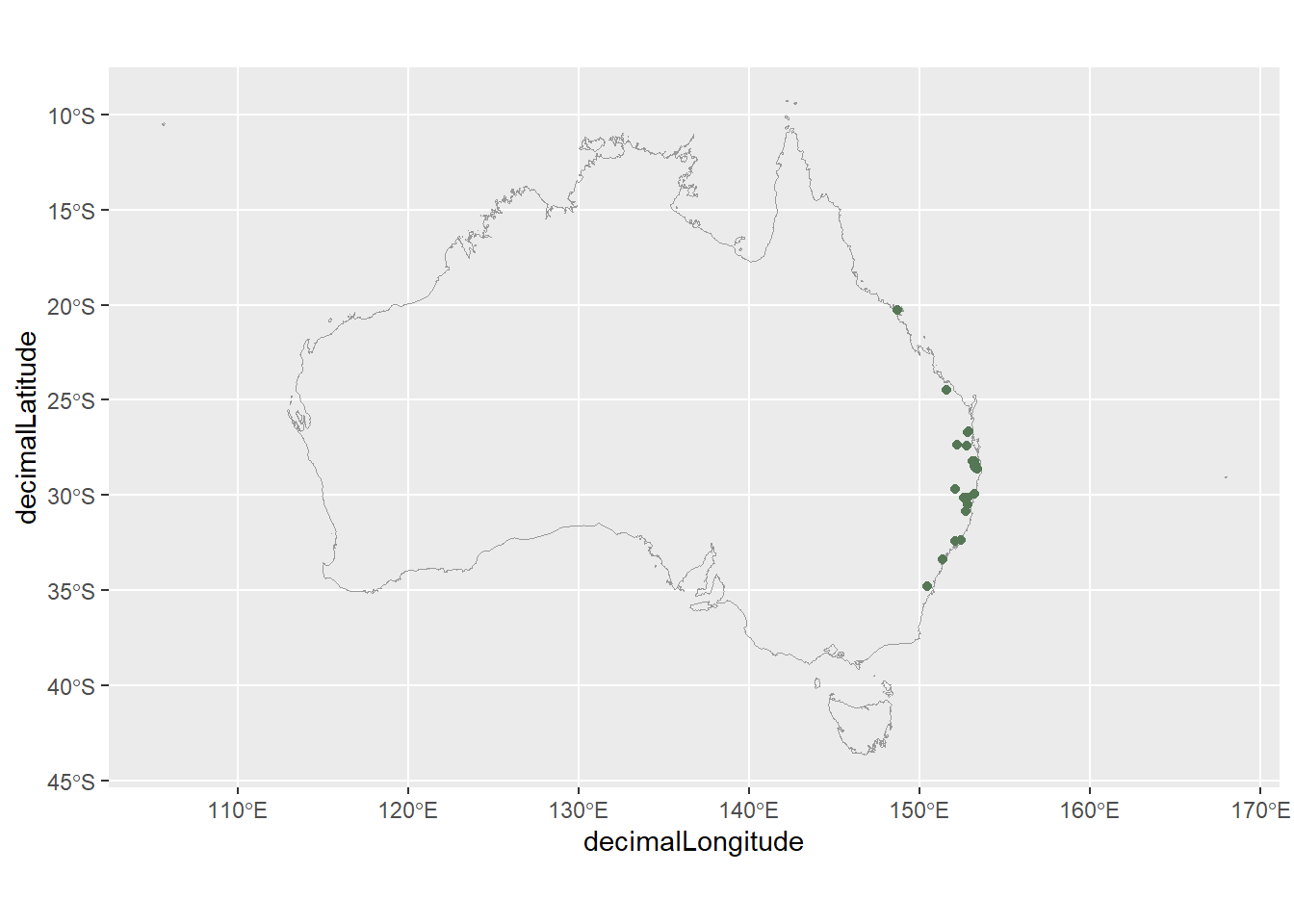
10.3 Issues to remove records
Some coordinates issues cannot be fixed or inferred. In this case, it is important that you identify which records have issues and remove them prior to analysis.
Here are some examples of geospatial errors that might need to be identified and removed in your dataset.
10.3.1 Flipped coordinates
Flipped coordinates typically appear as a clustering of points, whereby swapping the latitude and longitude will place the coordinates where they are expected (Jin and Yang 2020).
Let’s use occurrence data of Kowari (a native, carnivorous mouse species) as an example (downloaded at the start of the chapter). If using the galah package, you can add the COUNTRY_COORDINATE_MISMATCH assertion column to your download to find occurrence records flagged as with mismatching coordinates and country data.
native_mice <- native_mice |>
drop_na(decimalLongitude, decimalLatitude)
native_mice |>
select(COUNTRY_COORDINATE_MISMATCH, everything())# A tibble: 1,334 × 131
COUNTRY_COORDINATE_MISMATCH scientificName decimalLongitude decimalLatitude
<lgl> <chr> <dbl> <dbl>
1 TRUE Dasyuroides byr… 31.5 -54.2
2 FALSE Dasyuroides byr… 140. -27.0
3 FALSE Dasyuroides byr… 140. -27.0
4 FALSE Dasyuroides byr… 141. -24.2
5 FALSE Dasyuroides byr… 135. -25.9
6 FALSE Dasyuroides byr… 141. -23.8
7 FALSE Dasyuroides byr… 140. -27.0
8 FALSE Dasyuroides byr… 140. -23.7
9 TRUE Dasyuroides byr… 32.3 -56.8
10 FALSE Dasyuroides byr… 139. -25.6
# ℹ 1,324 more rows
# ℹ 127 more variables: eventDate <dttm>, country <chr>, countryCode <chr>,
# locality <chr>, AMBIGUOUS_COLLECTION <lgl>, AMBIGUOUS_INSTITUTION <lgl>,
# BASIS_OF_RECORD_INVALID <lgl>, biosecurityIssue <lgl>,
# COLLECTION_MATCH_FUZZY <lgl>, COLLECTION_MATCH_NONE <lgl>,
# CONTINENT_COORDINATE_MISMATCH <lgl>, CONTINENT_COUNTRY_MISMATCH <lgl>,
# CONTINENT_DERIVED_FROM_COORDINATES <lgl>, …Sometimes, flipped coordinates can be fixed by switching the latitude and longitude coordinates. Other times, like in this example, the way to fix the coordinates isn’t obvious.
ggplot() +
geom_sf(data = aus) +
geom_point(data = native_mice,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = COUNTRY_COORDINATE_MISMATCH)) +
pilot::scale_color_pilot()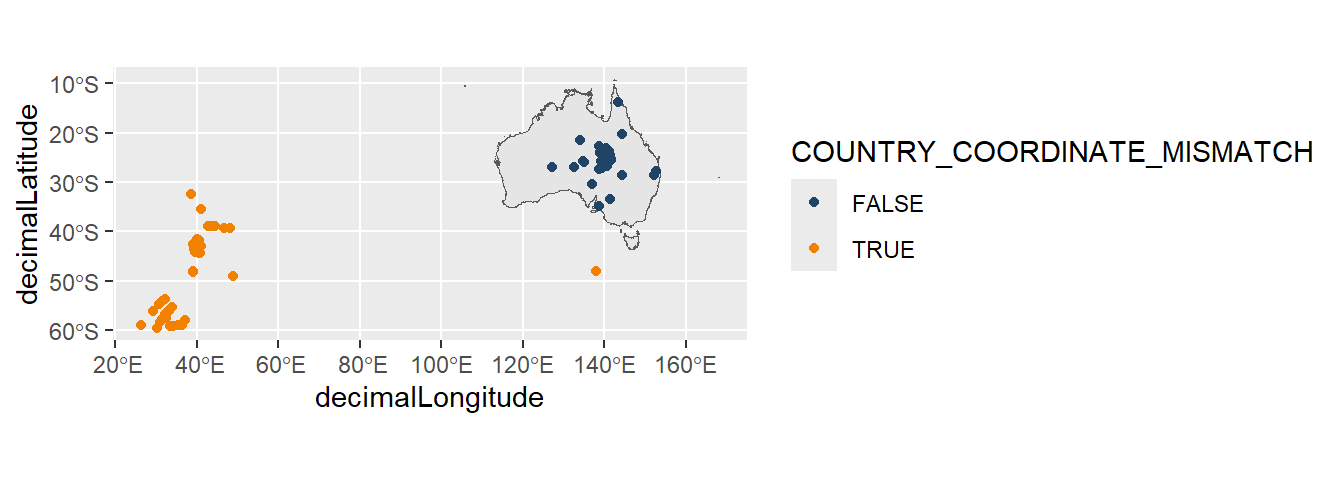
To remove these points from our data, we can filter our records to only records with coordinates within Australia’s land border.
native_mice_filtered <- native_mice |>
filter(decimalLongitude > 100,
decimalLongitude < 155,
decimalLatitude > -45,
decimalLatitude < -10)
ggplot() +
geom_sf(data = aus) +
geom_point(data = native_mice_filtered,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = COUNTRY_COORDINATE_MISMATCH)) +
pilot::scale_color_pilot()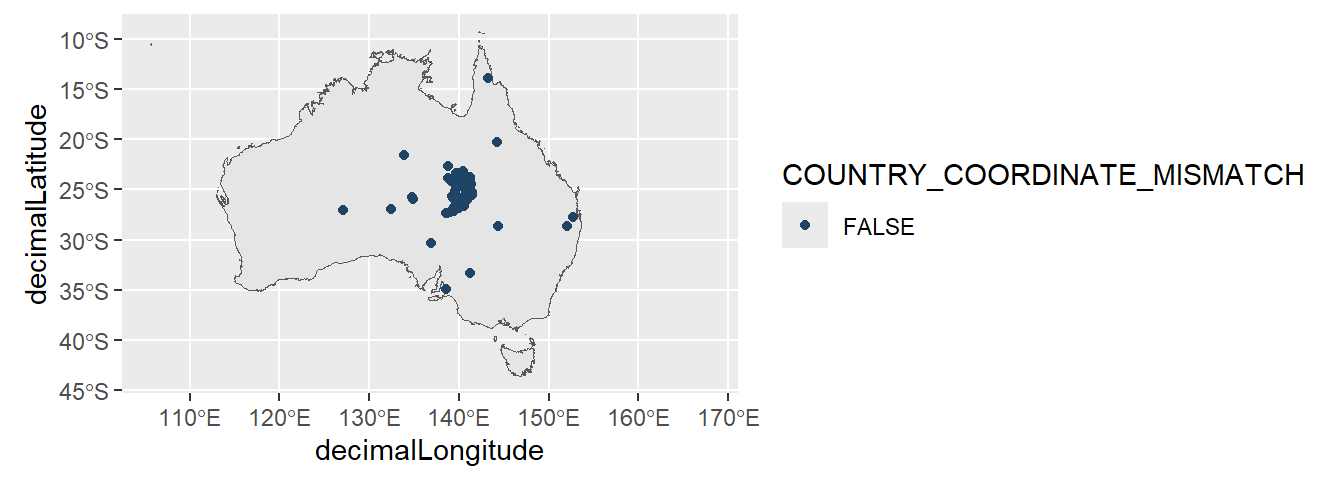
10.3.2 Zero coordinates
Some records are mistakenly recorded with zero as their latitude and/or longitude coordinates. These records will not accurately represent their valid location and must be removed.
Let’s use acacia data as an example (downloaded at the start of the chapter). If using the galah package, you can add the ZERO_COORDINATE assertion column to your download to find occurrence records flagged as suspicious for zero coordinates.
acacias <- acacias |>
drop_na(decimalLatitude, decimalLongitude) # remove NA values
acacias |>
select(ZERO_COORDINATE, everything())# A tibble: 10,704 × 11
ZERO_COORDINATE recordID scientificName taxonConceptID decimalLatitude
<lgl> <chr> <chr> <chr> <dbl>
1 FALSE 0013ae12-fda4-… Acacia aneura https://id.bi… -31.5
2 FALSE 00197d65-f235-… Acacia aneura https://id.bi… -29.5
3 FALSE 001a3cbb-a370-… Acacia aneura… https://id.bi… -28.0
4 FALSE 00238db7-6c4e-… Acacia aneura… https://id.bi… -34.1
5 FALSE 00276566-b590-… Acacia aneura https://id.bi… -29.7
6 FALSE 0029f3cf-b541-… Acacia aneura https://id.bi… -29.0
7 FALSE 0034f771-a3e1-… Acacia aneura https://id.bi… -31.1
8 FALSE 0035cb4f-85e5-… Acacia aneura… https://id.bi… -29.5
9 FALSE 003cce3b-f3f8-… Acacia aneura https://id.bi… -29.1
10 FALSE 0049bcbb-c8c2-… Acacia aneura https://id.bi… -25.5
# ℹ 10,694 more rows
# ℹ 6 more variables: decimalLongitude <dbl>, eventDate <dttm>,
# occurrenceStatus <chr>, dataResourceName <chr>, countryCode <chr>,
# locality <chr>We can see the suspicious record in orange on our map.
ggplot() +
geom_sf(data = aus) +
geom_point(data = acacias,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = ZERO_COORDINATE)) +
pilot::scale_color_pilot()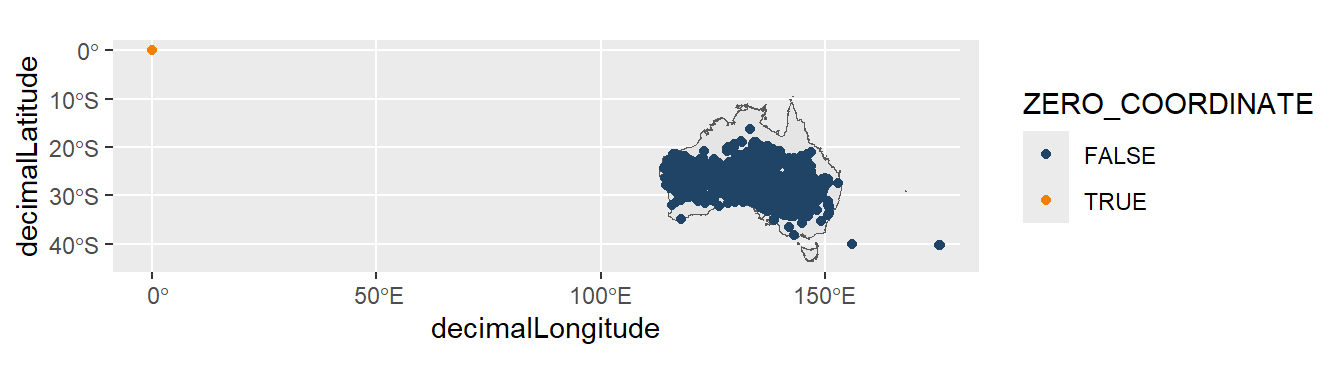
We can remove the problematic record by filtering our data to remove records with longitude or latitude coordinates that equal zero.
acacias_filtered <- acacias |>
filter(decimalLongitude != 0,
decimalLatitude != 0)
ggplot() +
geom_sf(data = aus) +
geom_point(data = acacias_filtered,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = ZERO_COORDINATE)) +
pilot::scale_color_pilot()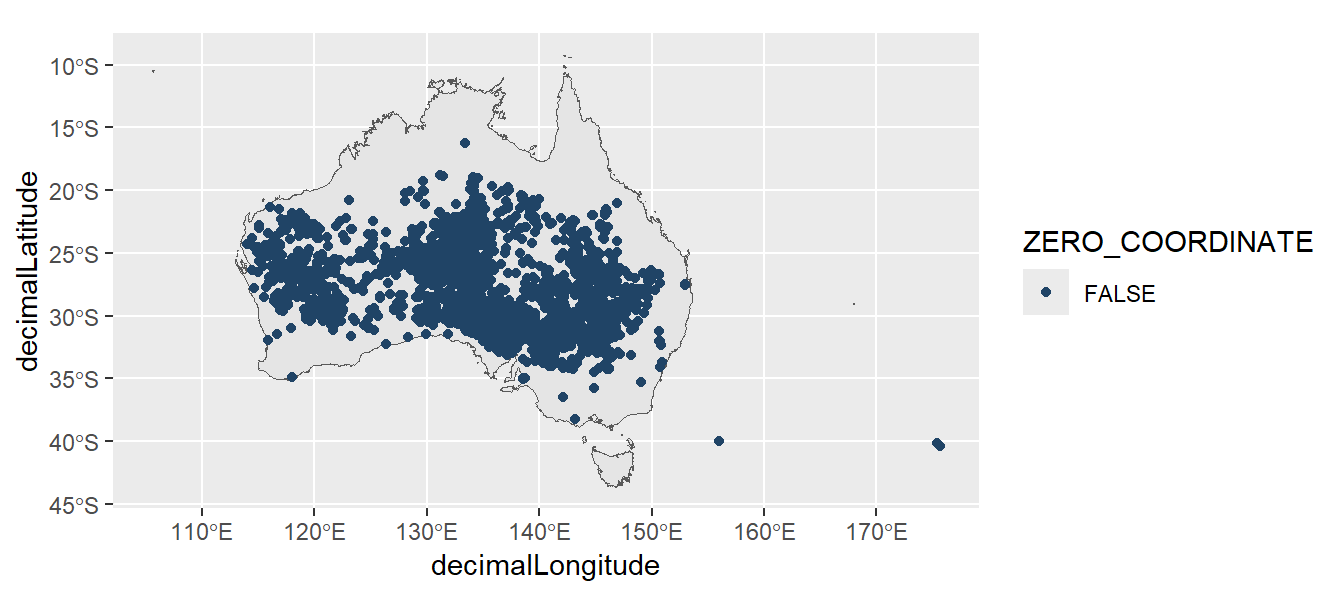
10.3.3 Centroids
Centroids, or coordinates that mark the exact centre point of an area, are sometimes assigned to an occurrence record when the original observation location was provided as a description. If a record was collected using a vague locality description or from incorrect georeferencing, centroids can be used to categorise the record into broadly the correct area1.
Let’s use common brown butterfly data for our example (downloaded at the start of the chapter). If using the galah package, we can add the COORDINATES_CENTRE_OF_COUNTRY or COORDINATES_CENTRE_OF_STATEPROVINCE assertions columns to your download to find occurrence records flagged as suspicious for centroid coordinates.
butterflies <- butterflies |>
drop_na(decimalLatitude, decimalLongitude) # remove NA values
butterflies |>
select(COORDINATES_CENTRE_OF_COUNTRY,
COORDINATES_CENTRE_OF_STATEPROVINCE,
everything())# A tibble: 335 × 12
COORDINATES_CENTRE_OF_COUNTRY COORDINATES_CENTRE_OF…¹ recordID scientificName
<lgl> <lgl> <chr> <chr>
1 FALSE FALSE 018f5a5… Heteronympha …
2 FALSE FALSE 02eef43… Heteronympha …
3 FALSE FALSE 03b39bb… Heteronympha …
4 FALSE FALSE 04c2ac2… Heteronympha …
5 FALSE FALSE 05cced1… Heteronympha …
6 FALSE FALSE 05ceb8b… Heteronympha …
7 FALSE FALSE 06679b8… Heteronympha …
8 FALSE FALSE 0704e7b… Heteronympha …
9 FALSE FALSE 0756bd4… Heteronympha …
10 FALSE FALSE 0774f2e… Heteronympha …
# ℹ 325 more rows
# ℹ abbreviated name: ¹COORDINATES_CENTRE_OF_STATEPROVINCE
# ℹ 8 more variables: taxonConceptID <chr>, decimalLatitude <dbl>,
# decimalLongitude <dbl>, eventDate <dttm>, occurrenceStatus <chr>,
# dataResourceName <chr>, countryCode <chr>, locality <chr>Filtering our data to records flagged as suspicious, we return one record.
butterflies |>
filter(
COORDINATES_CENTRE_OF_COUNTRY == TRUE |
COORDINATES_CENTRE_OF_STATEPROVINCE == TRUE
)# A tibble: 1 × 12
recordID scientificName taxonConceptID decimalLatitude decimalLongitude
<chr> <chr> <chr> <dbl> <dbl>
1 89186e67-be72-… Heteronympha … https://biodi… -31.3 147.
# ℹ 7 more variables: eventDate <dttm>, occurrenceStatus <chr>,
# dataResourceName <chr>, countryCode <chr>, locality <chr>,
# COORDINATES_CENTRE_OF_COUNTRY <lgl>,
# COORDINATES_CENTRE_OF_STATEPROVINCE <lgl>The suspicious record is the single orange point on our map.
ggplot() +
geom_sf(data = aus) +
geom_point(data = butterflies,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = COORDINATES_CENTRE_OF_STATEPROVINCE)) +
pilot::scale_color_pilot() +
theme(legend.position = "none")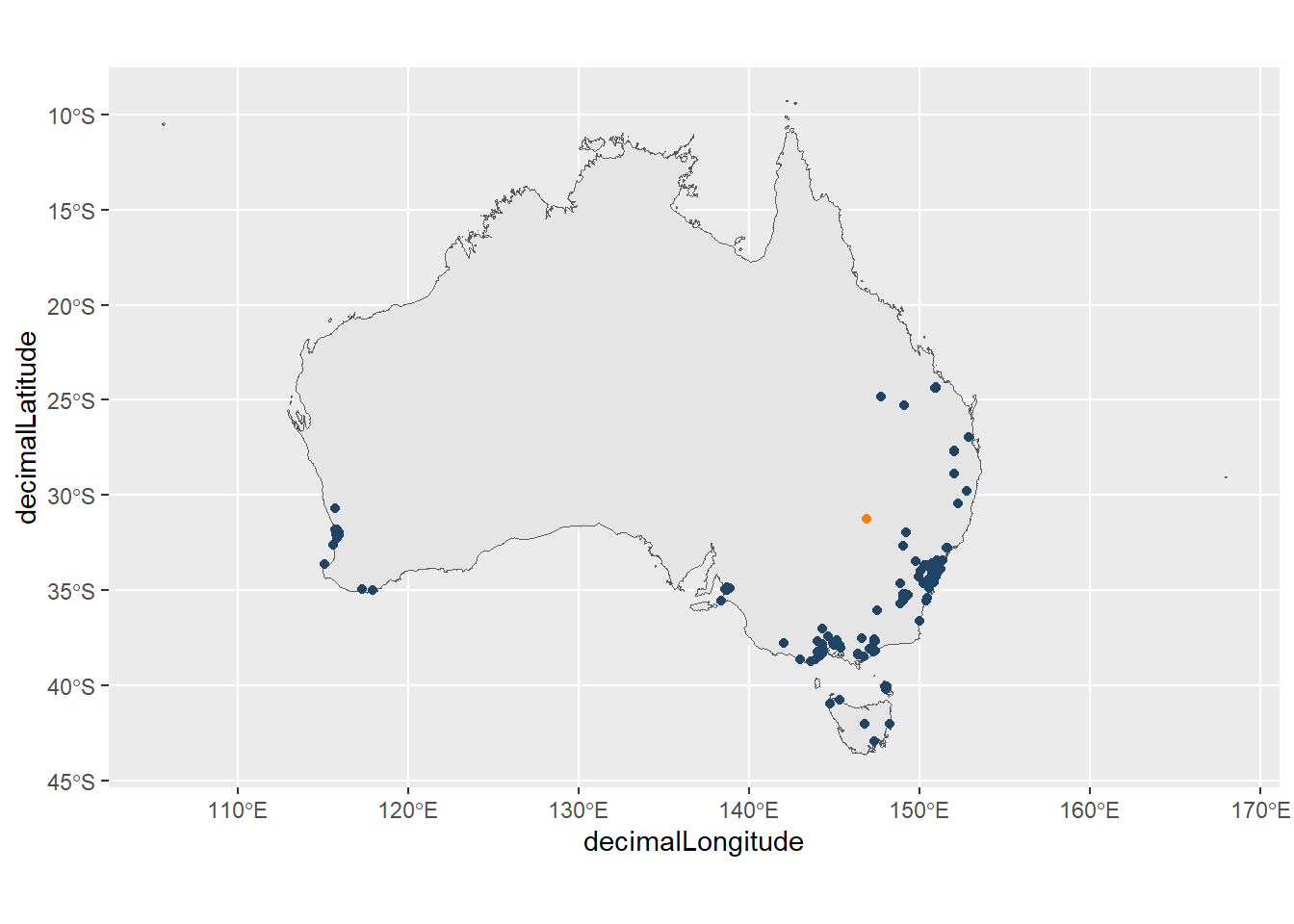
We can remove this data point by excluding this record from our dataframe.
butterflies_filtered <- butterflies |>
filter(COORDINATES_CENTRE_OF_STATEPROVINCE == FALSE)
ggplot() +
geom_sf(data = aus) +
geom_point(data = butterflies_filtered,
aes(x = decimalLongitude,
y = decimalLatitude,
colour = COORDINATES_CENTRE_OF_STATEPROVINCE)) +
pilot::scale_color_pilot() +
theme(legend.position = "none")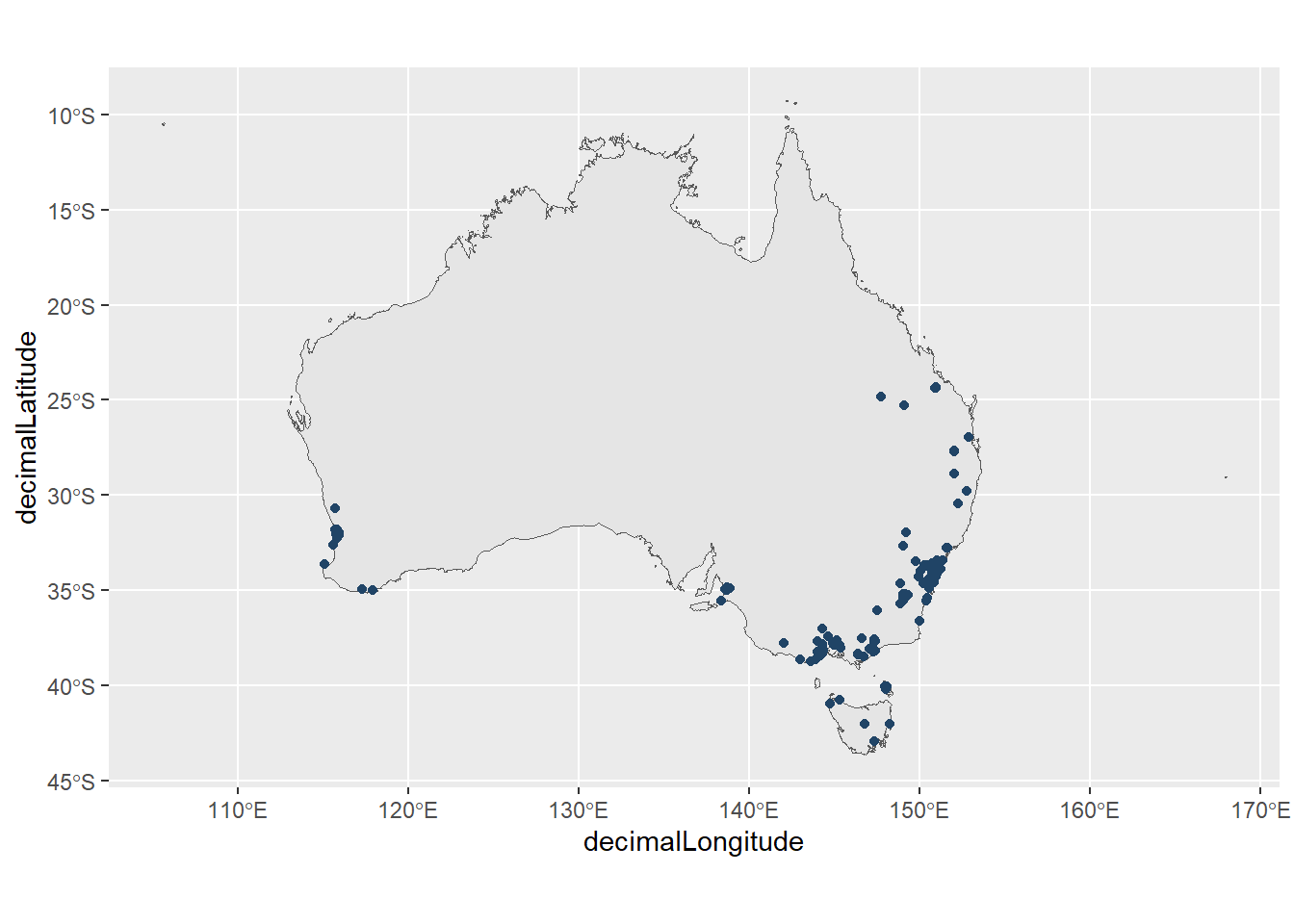
10.3.4 Cities, zoos, aquariums, museums & herbaria
Some observations are recorded in locations where animals and plants live but do not naturally occur. A common example is observations recorded at public facilities like zoos, aquariums and botanic gardens.
Other times, observations are recorded in places where specimens of animals and plants might be stored, but not where they were observed. Common examples are museums and herbaria.
In some cases, like with records of the Gorse Bitter-pea (downloaded at the start of the chapter), these locations can appear suspicious but not overly obvious. When we map these observations, there is a tailing distribution of points in Western Australia with several points located near the west coast of Australia.
bitter_peas <- bitter_peas |>
drop_na(decimalLongitude, decimalLatitude) # remove NA values
ggplot() +
geom_sf(data = aus) +
geom_point(data = bitter_peas,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = pilot::pilot_color("navy"))
Suspiciously, if we Google the coordinates of the Western Australia Herbarium, the coordinates overlap with one of the points. We have highlighted this point in orange.
ggplot() +
geom_sf(data = aus) +
geom_point(data = bitter_peas,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = "#204466") +
geom_point(aes(x = 115.8, y = -31.9), # point coordinates
colour = "#f28100") +
theme(legend.position = "none")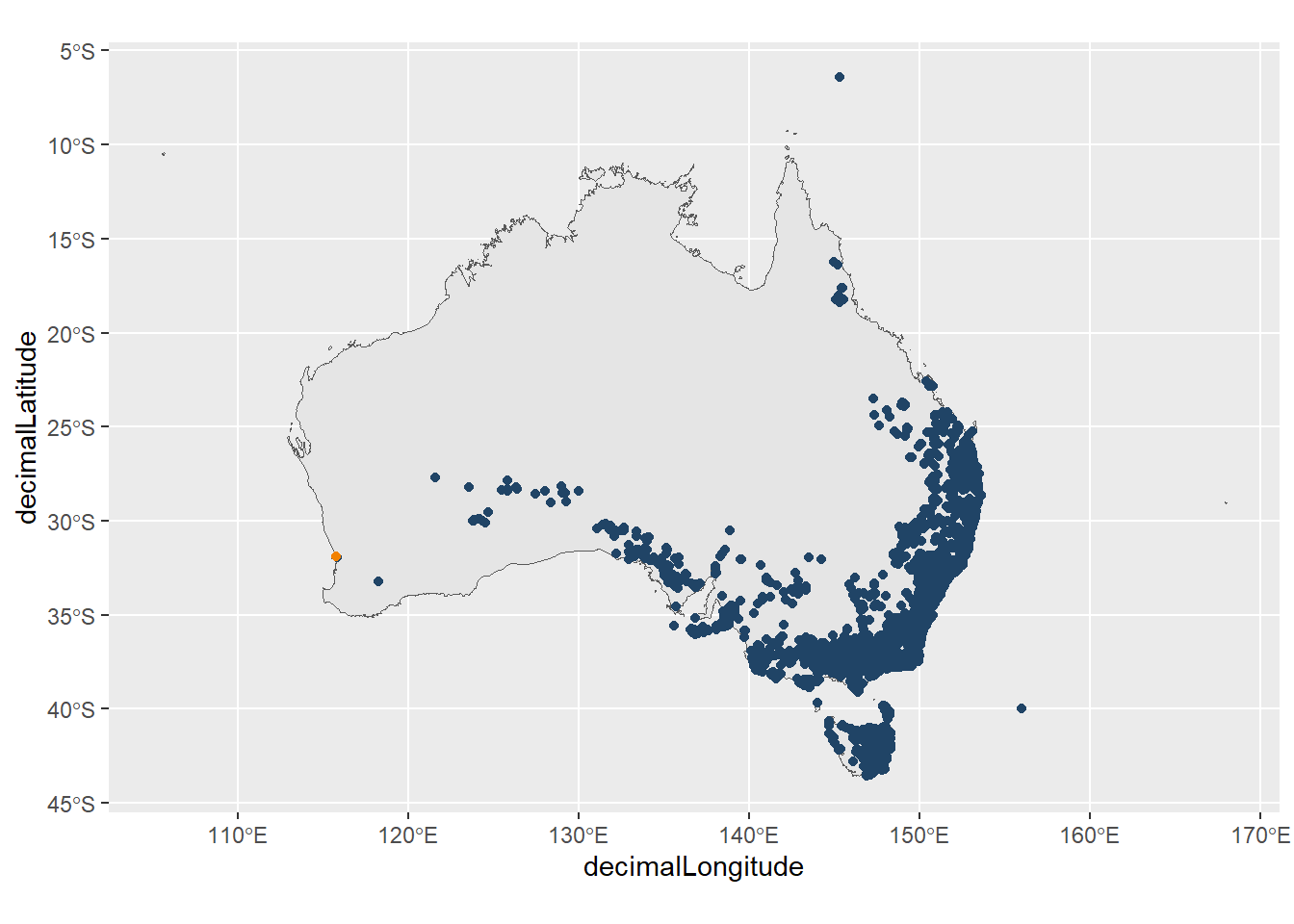
Filtering our data to the two left-most data points reveals that the data resources that supplied those records are both state herbaria.
bitter_peas |>
filter(decimalLongitude < 120) |>
select(dataResourceName)# A tibble: 2 × 1
dataResourceName
<chr>
1 National Herbarium of Victoria (MEL) AVH data
2 NSW AVH feed Having identified they could be can remove these records from our data.
bitter_peas_filtered <- bitter_peas |>
filter(decimalLongitude > 120)
ggplot() +
geom_sf(data = aus) +
geom_point(data = bitter_peas_filtered,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = "#204466")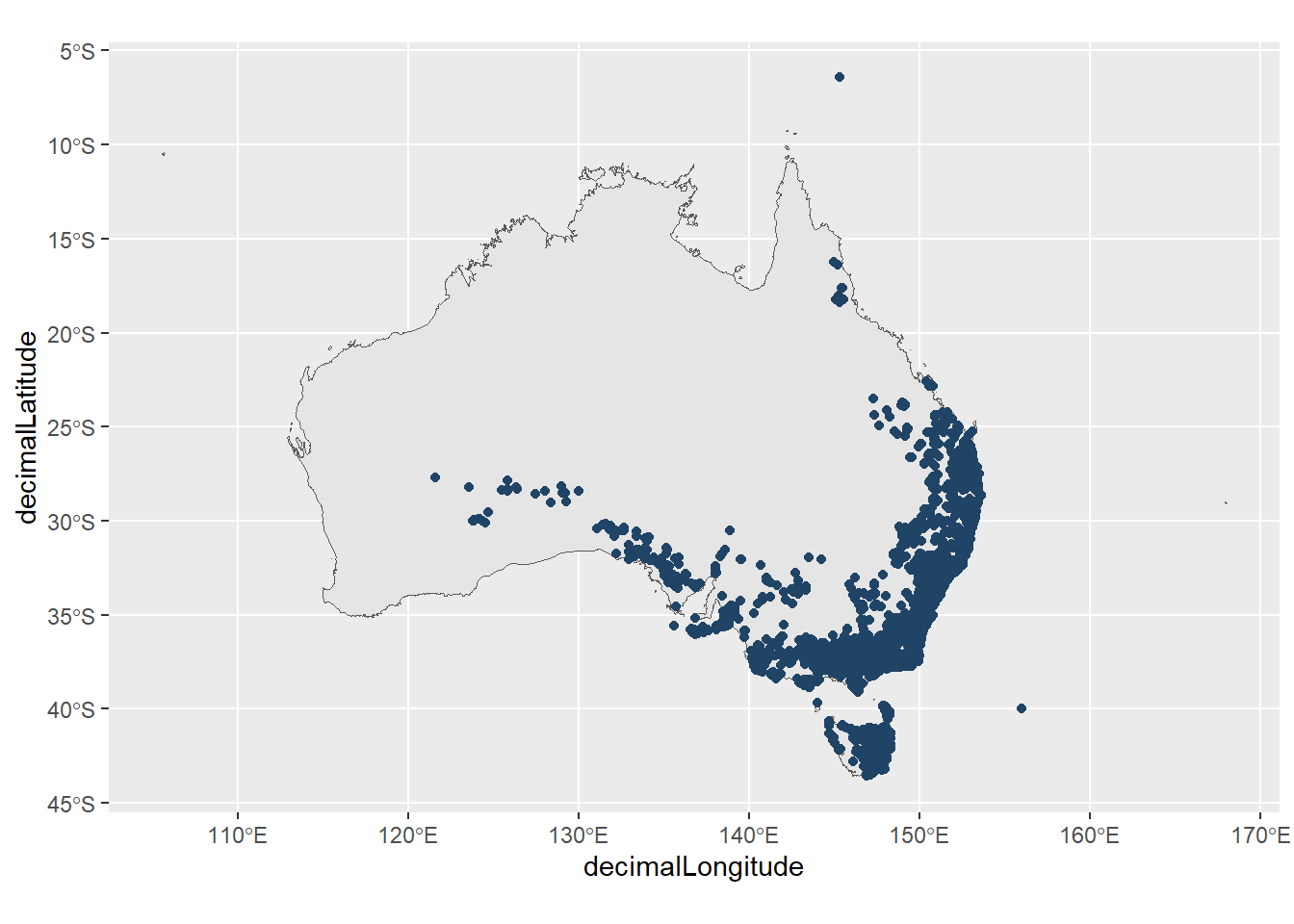
basisOfRecord
You can use the field basisOfRecord to avoid including records from museums and herbaria when creating your query in galah.
library(galah)
# Show values in `basisOfRecord` field
search_all(fields, "basisOfRecord") |>
show_values()
# Filter basis of record to only human observations
galah_call() |>
identify("Daviesia ulicifolia") |>
filter(basisOfRecord == "HUMAN_OBSERVATION") |>
atlas_counts()10.4 Use expert distributions
Coming soon…
10.5 Packages
CoordinateCleaner
The CoordinateCleaner package is a package for automated flagging of common spatial and temporal errors of biological and paleaontological data. It is particularly good at cleaning data from GBIF.
Here is an example of a general cleaning function, but there are many more bespoke options that the package offers.
library(CoordinateCleaner)
# Run record-level tests
coordinate_tests <- clean_coordinates(x = butterflies,
species = "scientificName")Reading layer `ne_50m_land' from data source
`C:\Users\KEL329\AppData\Local\Temp\Rtmpqe6Xq1\ne_50m_land.shp'
using driver `ESRI Shapefile'
Simple feature collection with 1420 features and 3 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -180 ymin: -89.99893 xmax: 180 ymax: 83.59961
Geodetic CRS: WGS 84summary(coordinate_tests) .val .equ .zer .cap .cen .sea .otl .gbf
0 0 0 22 0 11 60 0
.inst .summary
13 95 plot(coordinate_tests)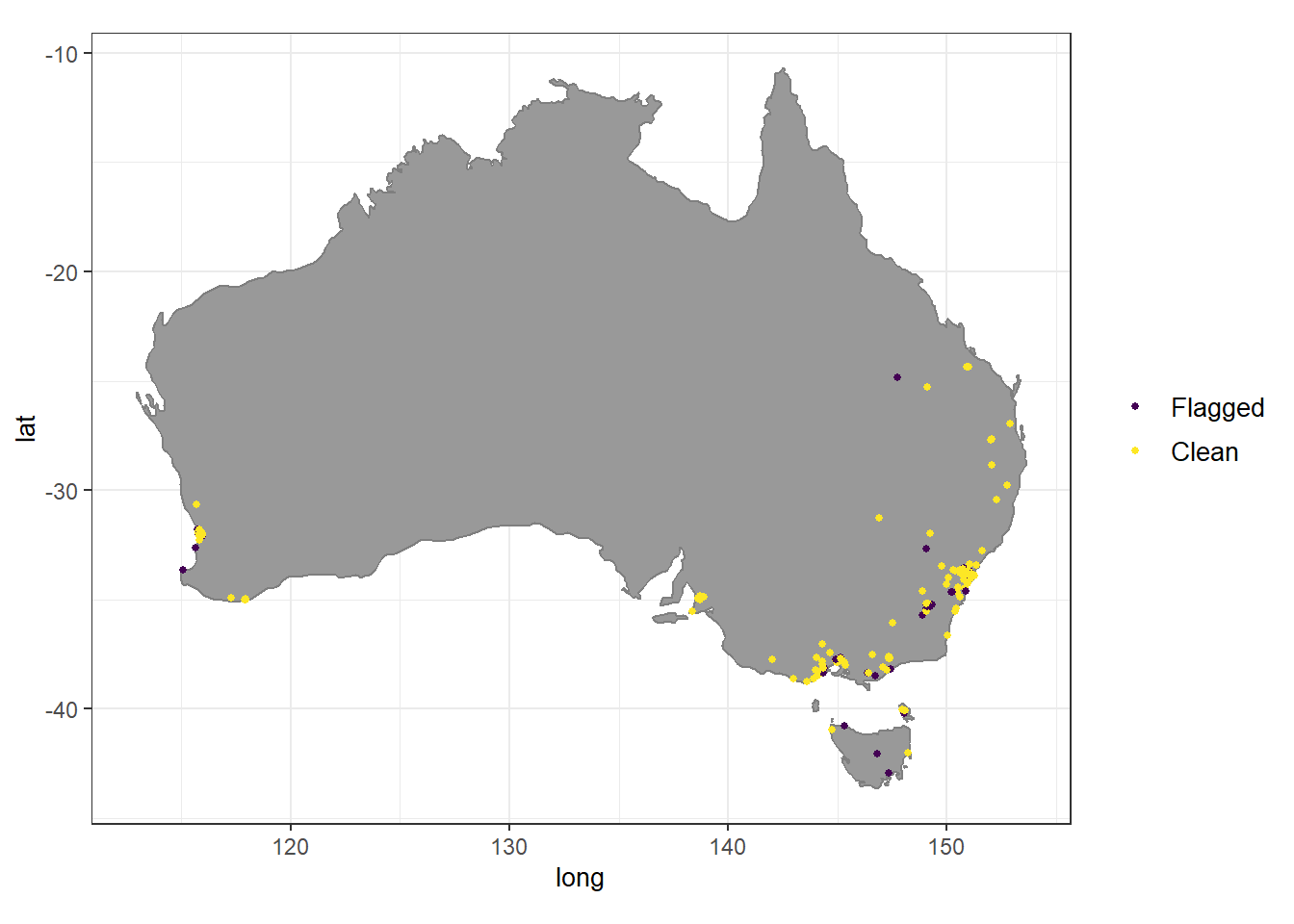
10.6 Summary
Each of the cleaning steps in this chapter do not have to be run in order, or even at all. Whether they are used is context dependent and taxon dependent. As an example, what is one species that has many “wrong” coordinates based on many of the steps listed above?
The Great White Shark.
Code
# Download occurrence records
sharks <- galah_call() |>
identify("Carcharodon carcharias") |>
filter(basisOfRecord == "HUMAN_OBSERVATION") |>
apply_profile(ALA) |>
atlas_occurrences()
# Retrieve map of Australia
aus <- st_transform(ozmap_country, 4326)
# Map occurrences
sharks |>
drop_na(decimalLongitude, decimalLatitude) |>
ggplot() +
geom_sf(data = aus,
colour = "grey60",
fill = "white") +
geom_point(data = sharks,
aes(x = decimalLongitude,
y = decimalLatitude),
colour = "#135277") +
theme_light()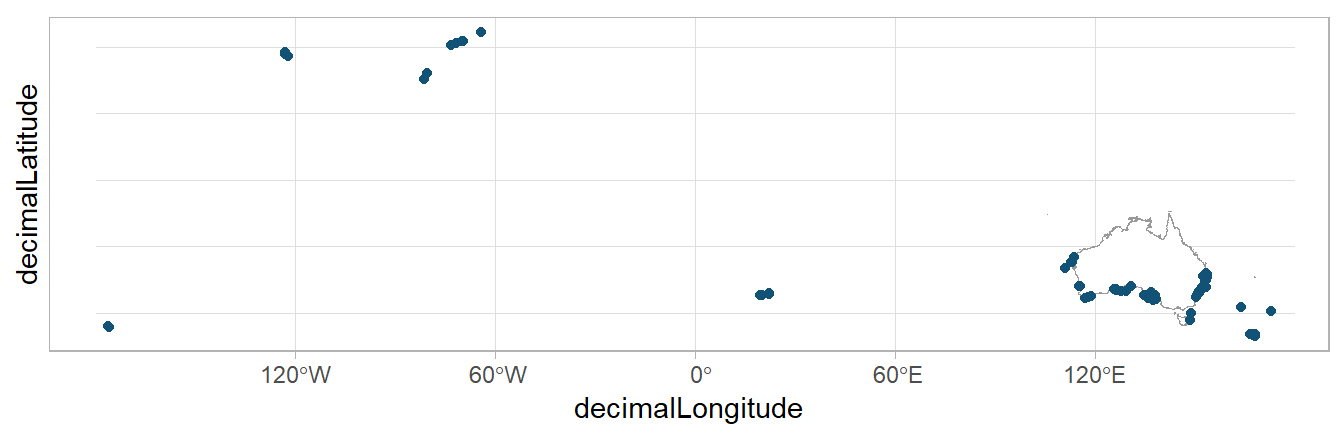
The difficulty with cleaning Great White Shark occurrence data is that these sharks have a massive habitat range, and these locations along (what appear to be) the North American coast and Madagascar could very well be true occurrences. In this case, we might need to decide the spatial range that makes sense for our specific investigation. Be sure to consider the taxonomic and spatial range of your species before jumping into data cleaning!
This can happen when record locations is incorrectly given as the physical location of the specimen, or because they represent individuals from captivity or grown in horticulture (but were not clearly labelled as such).↩︎







